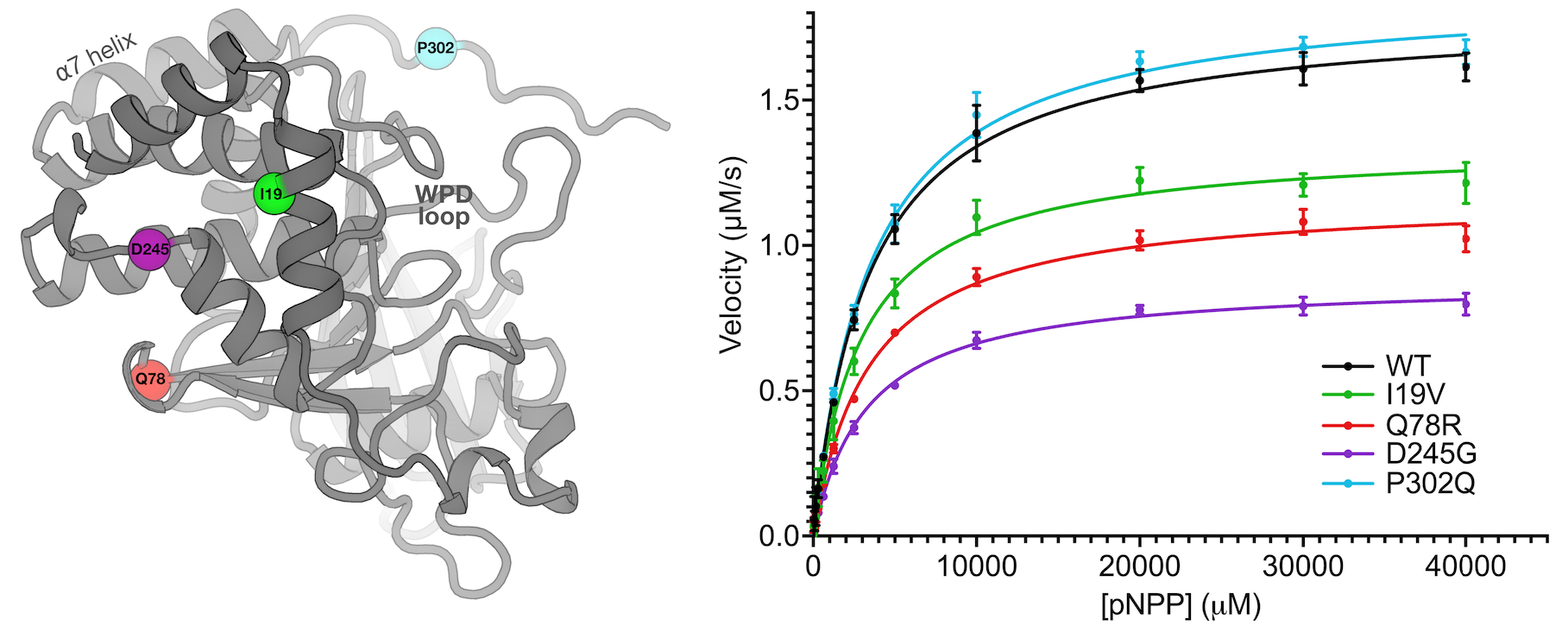
Perdikari A*, Woods VA*, Ebrahim A*, Lawler K, Bounds R, Shah DS, Singh NI, Mehlman (Skaist) T, Riley BT, Sharma S, Morris JW, Keogh JM, Henning E, Smith M, Farooqi IS**, DA Keedy** (** co-corresponding authors). Structures of human protein tyrosine phosphatase variants reveal targetable allosteric sites. Journal of Biological Chemistry. 2025.
- PMID: 41135676
- bioRxiv Preprint: 2024.08.05.603709
- Full Text
- Deposited Structures: 9CYO, 9CYP, 9CYQ, 9CYR
- Farooqi group at Cambridge
- STILTS - Study Into Lean and Thin Subjects
- Beamline 12-1 at SSRL
- Remote room-temperature crystallography at SSRL
- X-ray diffraction data at Zenodo
Cavender CE, Case DA, Chen JCH, Chong LT, Keedy DA, Lindorff-Larsen K, Mobley DL, Ollila OHS, Oostenbrink C, Robustelli P, Voelz VA, Wall ME, Wych DC, Gilson MK. Structure-Based Experimental Datasets for Benchmarking Protein Simulation Force Fields. LiveCoMS. 2025.

- PMID:
- Full Text
- Preprint on arXiv
- Michael Gilson
- Open Force Field Initiative
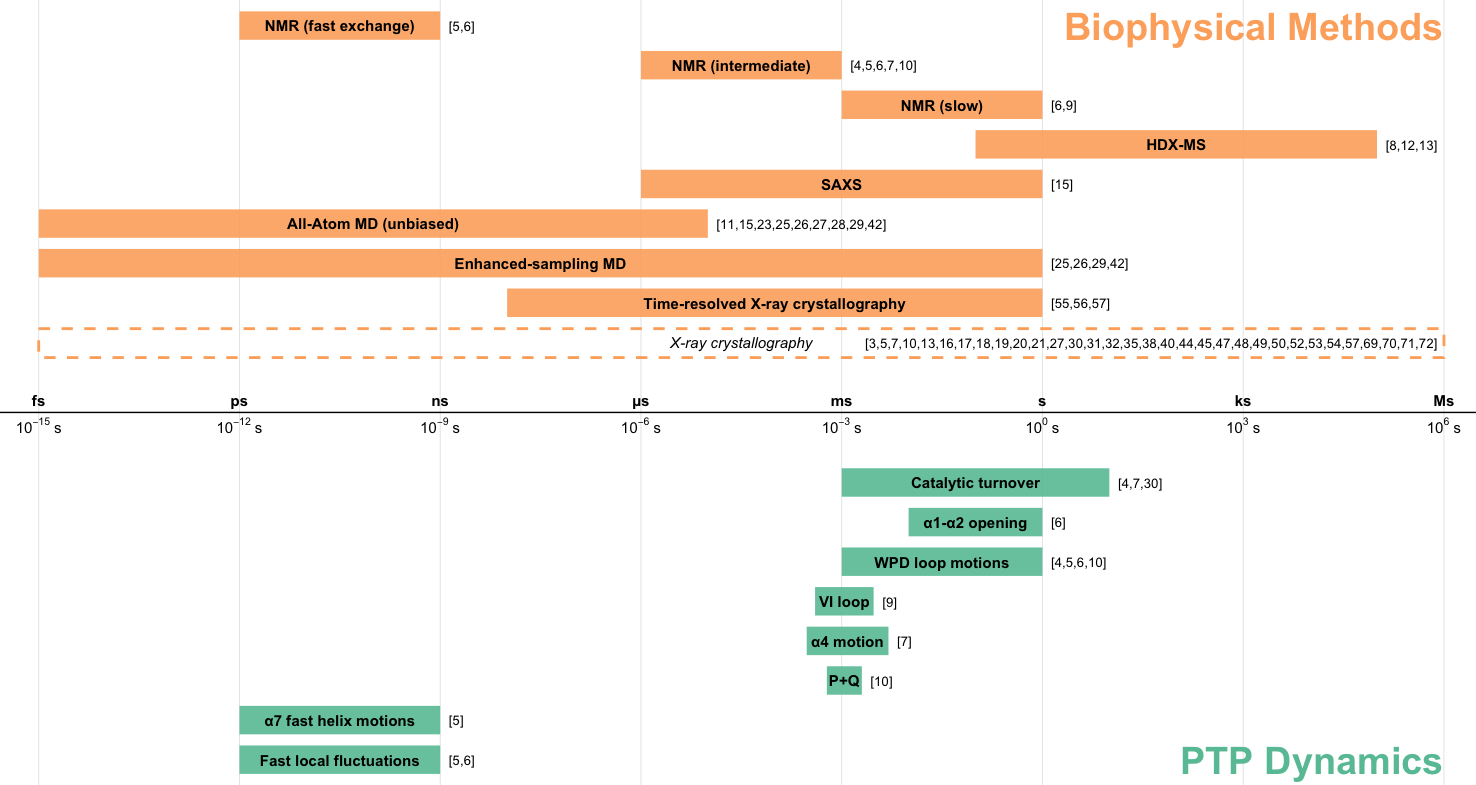
Woods VA*, Sharma S*, Lemberikman AM, Keedy DA. Orchestrating function: concerted dynamics, allostery, and catalysis in protein tyrosine phosphatases. Current Opinion in Structural Biology. 2025.
- PMID: 40752367
- PMCID: PMC12321194
- Full Text
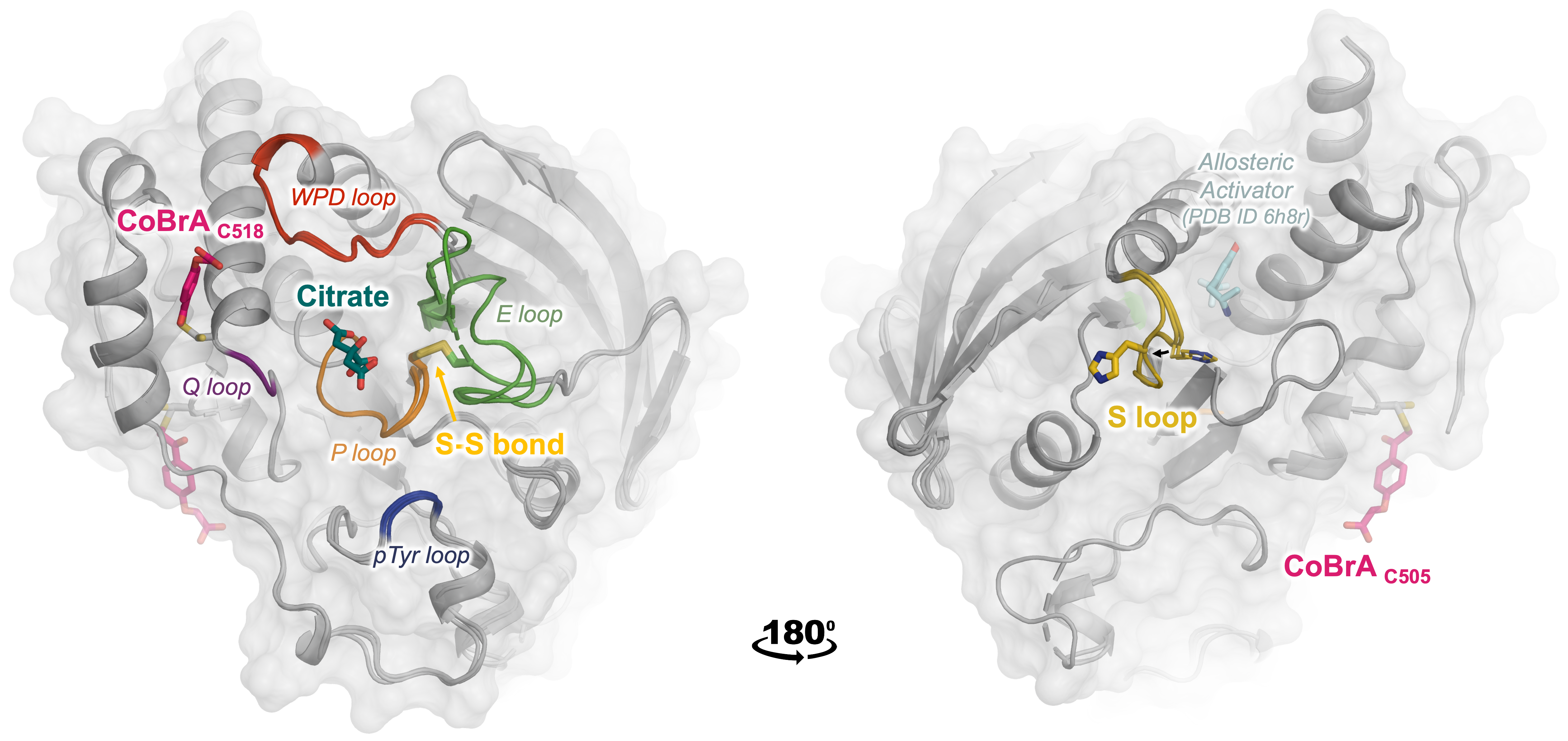
Guerrero L, Ebrahim A, Riley BT, Kim SH, Bishop AC, Wu J, Han YN, Tautz L, Keedy DA. Three STEPs Forward: A Trio of Unexpected Structures of PTPN5. Proteins: Structure, Function, and Bioinformatics. 2025.
- PMID: 40616465
- PMCID: PMC12270549
- bioRxiv Preprint: 2024.11.20.624168
- Full Text
- Deposited Structures: 9EEX, 9EEY, 9EEZ
- Bishop group at Amherst
- Tautz group at Sanford Burnham
- NYX beamline at NSLS-II
- AMX beamline at NSLS-II
- FlexX beamline at MacCHESS
- X-ray diffraction data at ProteinDiffraction.org for 9EEX
- X-ray diffraction data at ProteinDiffraction.org for 9EEY
- X-ray diffraction data at ProteinDiffraction.org for 9EEZ
- Sneak Peek during peer review at Structure
Raju A, Sharma S, Riley BT, Djuraev S, Tan Y, Kim M, Mahmud T, Keedy DA. Mapping allosteric rewiring in related protein structures from collections of crystallographic multiconformer models. bioRxiv preprint. 2025.
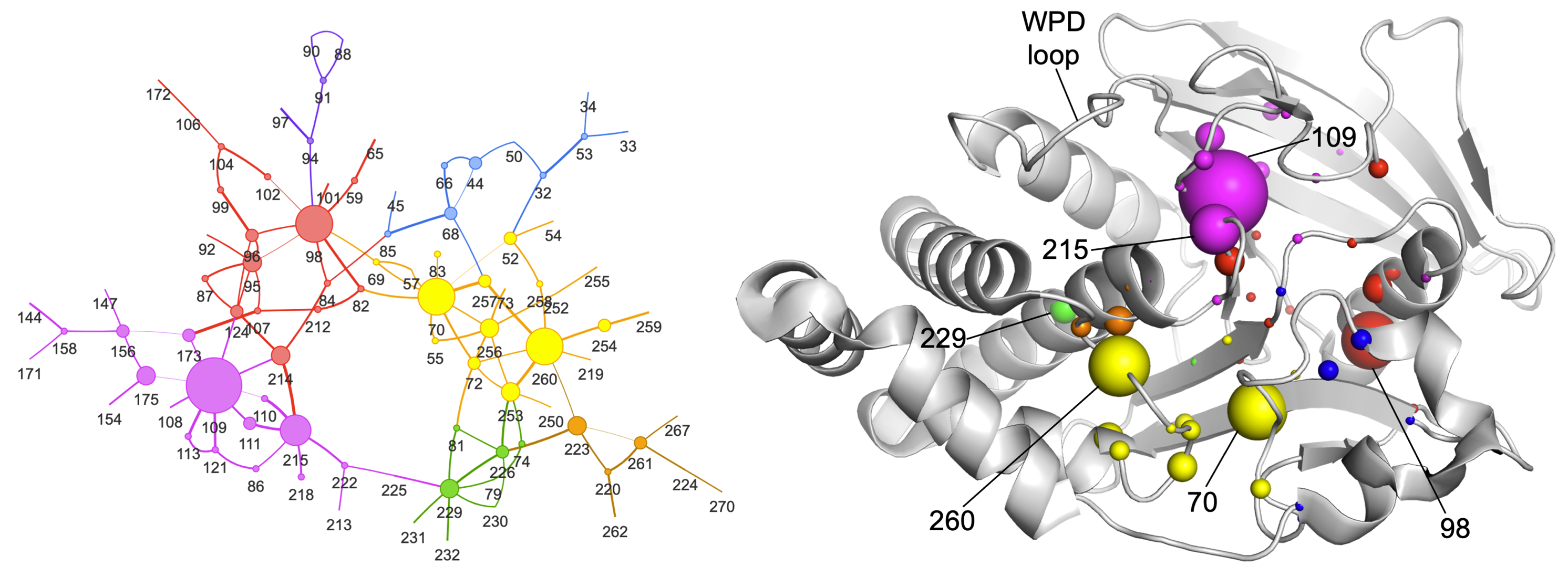
- PMID:
- bioRxiv Preprint: 2025.05.23.655529
- Full Text
- RINFAIRE code on GitHub
- Supplementary data on Zenodo
- qFit code on GitHub
- Bluesky thread
Wu J, Baranowski MR, Aleshin AE, Isiorho EA, Lambert LJ, De Backer LJS, Han YN, Das R, Sheffler DJ, Bobkov AA, Lemberikman AM, Keedy DA, Cosford NDP, Tautz L. Fragment Screening Identifies Novel Allosteric Binders and Binding Sites in the VHR (DUSP3) Phosphatase. ACS Omega. 2025.
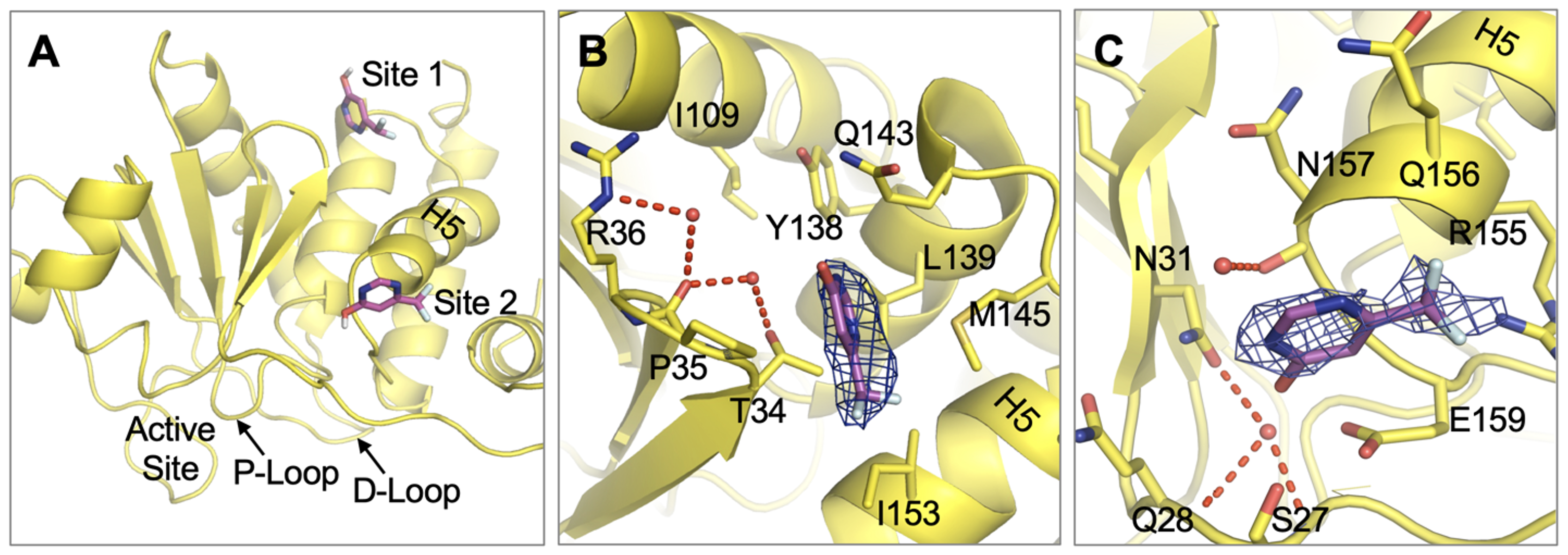
- PMID: 39959108
- PMCID: PMC11822521
- Full Text
- Deposited Structures: 8TK2, 8TK3, 8TK4, 8TK5, 8TK6, 9DJ9
- Tautz group at Sanford Burnham
Chartier CA, Woods VA, Xu Y, van Vlimmeren AE, Jovanovic M, McDermott AE, Keedy DA, Shah NH. Allosteric regulation of the tyrosine phosphatase PTP1B by a protein-protein interaction. Protein Science. 2025.
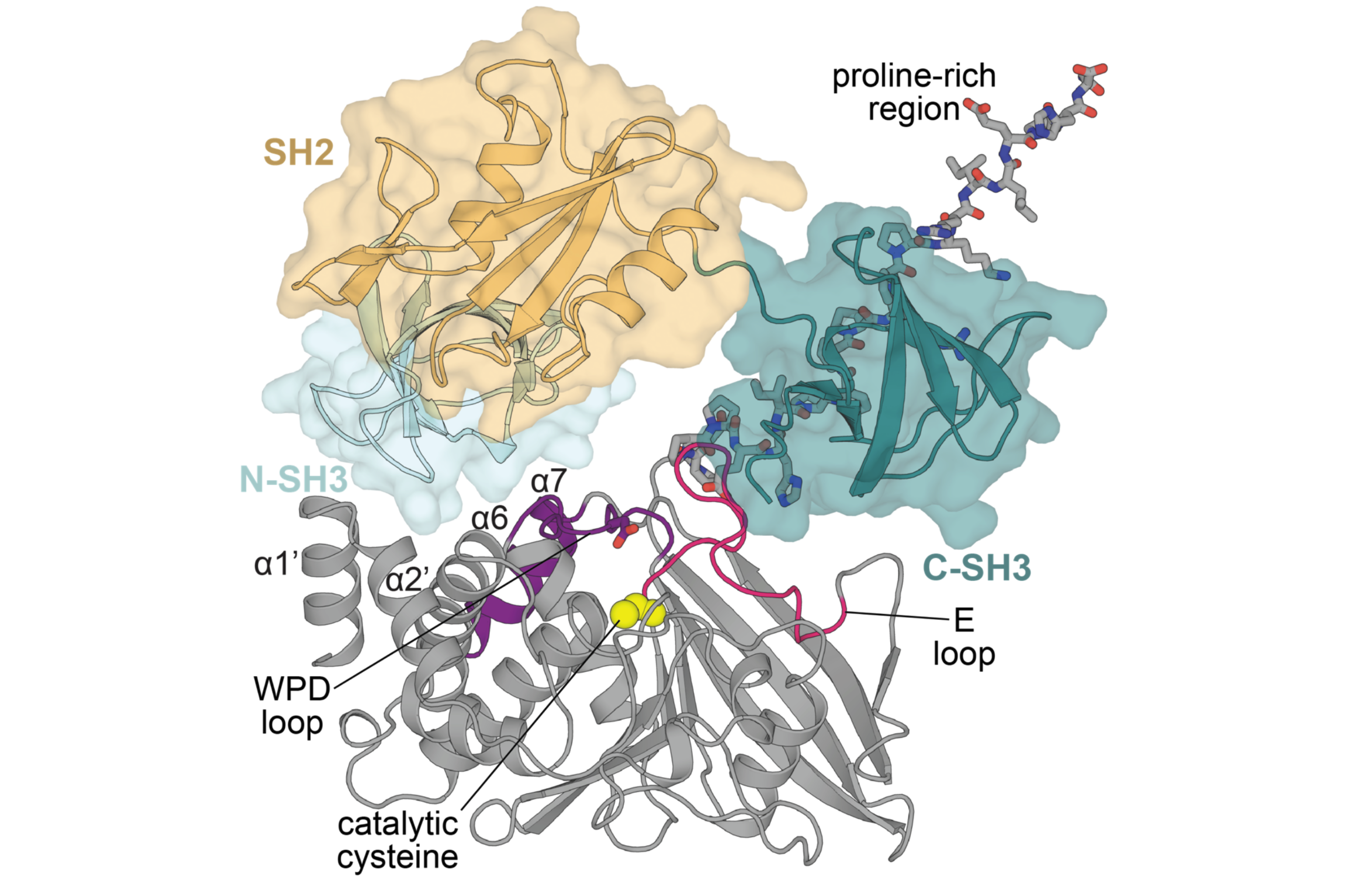
- PMID: 39723820
- PMCID: PMC11670308
- bioRxiv Preprint: 2024.07.16.603632
- Full Text
- Shah lab at Columbia
Wankowicz SA, Ravikumar A, Sharma S, Riley BT, Raju A, Hogan D, van den Bedem H, Keedy DA, Fraser JS. Automated multiconformer model building for X-ray crystallography and cryo-EM. eLife. 2024.
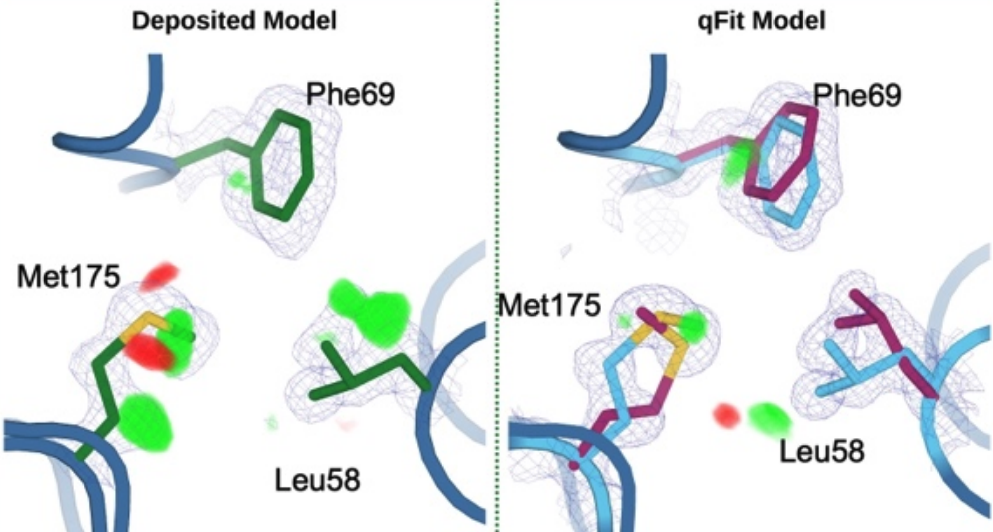
- PMID: 38904665
- PMCID: PMC11192534
- bioRxiv Preprint: 2023.06.28.546963
- Full Text
- qFit code on GitHub
- qFit application on SBGrid
- Fraser lab at UCSF
Mehlman (Skaist) T, Ginn HM, Keedy DA. An expanded trove of fragment-bound structures for the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data. Structure. 2024.
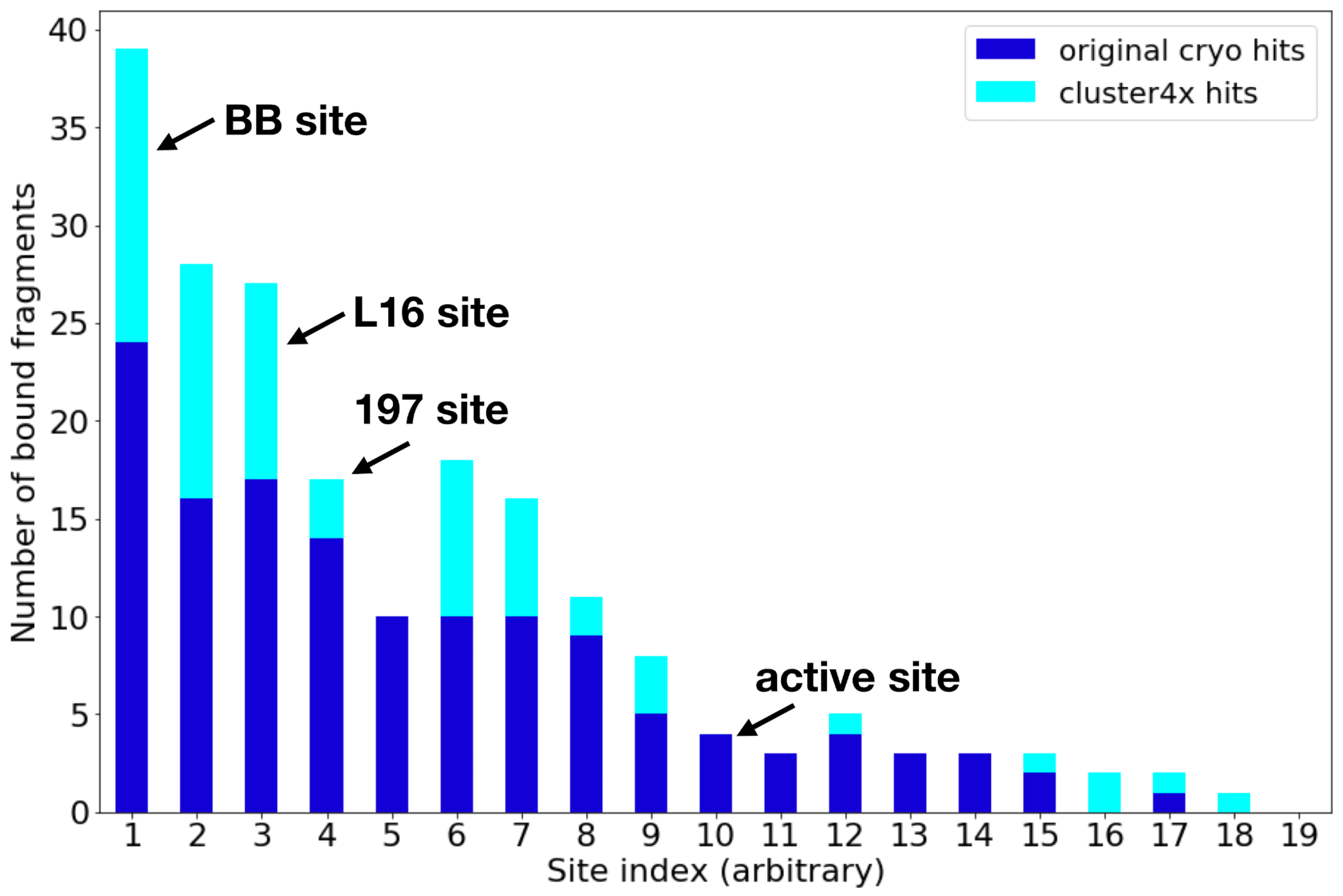
- PMID: 38861991
- PMCID: PMC11316629
- bioRxiv Preprint: 2024.01.05.574428
- Full Text
- Deposited Structures: 7GS7, 7GS8, 7GS9, 7GSA, 7GSB, 7GSC, 7GSD, 7GSE, 7GSF, 7GSG, 7GSH, 7GSI, 7GSJ, 7GSK, 7GSL, 7GSM, 7GSN, 7GSO, 7GSQ, 7GSR, 7GST, 7GSU, 7GSV, 7GSW, 7GSX, 7GSY, 7GSZ, 7GT0, 7GT1, 7GT2, 7GT3, 7GT4, 7GT5, 7GT6, 7GT7, 7GT8, 7GT9, 7GTA, 7GTB, 7GTC, 7GTD, 7GTE, 7GTF, 7GTG, 7GTH, 7GTI, 7GTJ, 7GTK, 7GTL, 7GTM, 7GTN, 7GTO, 7GTP, 7GTQ, 7GTR, 7GTS, 7GTT, 7GTU, 7GTV, 7GTW, 7GTX, 7GTY, 7GTZ, 7GU0, 7GU1, 7GU2, 7GU3, 7GU4, 7GU5, 7GU6, 7GU7, 7GU8, 7GU9, 7GUA, 7GUB, 7GUC
- Protein Machinists lab - Helen Ginn
- cluster4x - Vagabond
- Datasets on Zenodo
- CUNY ASRC news post
Woods VA, Abzalimov RR, Keedy DA. Native dynamics and allosteric responses in PTP1B probed by high-resolution HDX-MS. Protein Science. 2024.
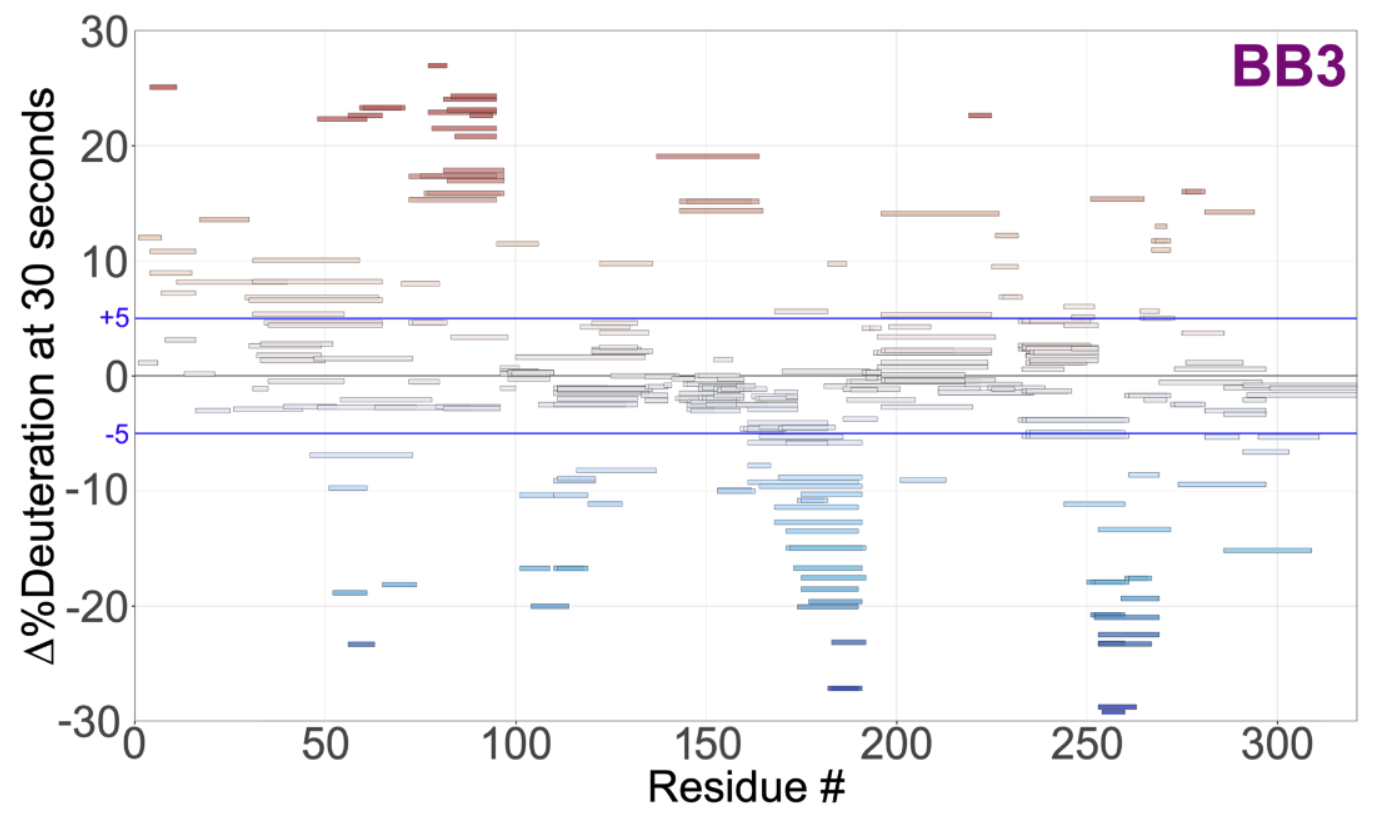
- PMID: 38801229
- PMCID: PMC11129624
- bioRxiv Preprint: 2023.07.12.548582
- Full Text
- ASRC SBI Mass Spectrometry Facility
- Twitter thread (v1)
- Twitter thread (v2)
Wallach I, …, Keedy DA, …, Smith KS, …, Singh N, …, Hossain S, …, Dzhumaev S, …, Azeem SM, …, Mehlman T, …, Woods VA…, Heifets A (688 total authors). AI is a viable alternative to high throughput screening: a 318-target study. Scientific Reports. 2024.
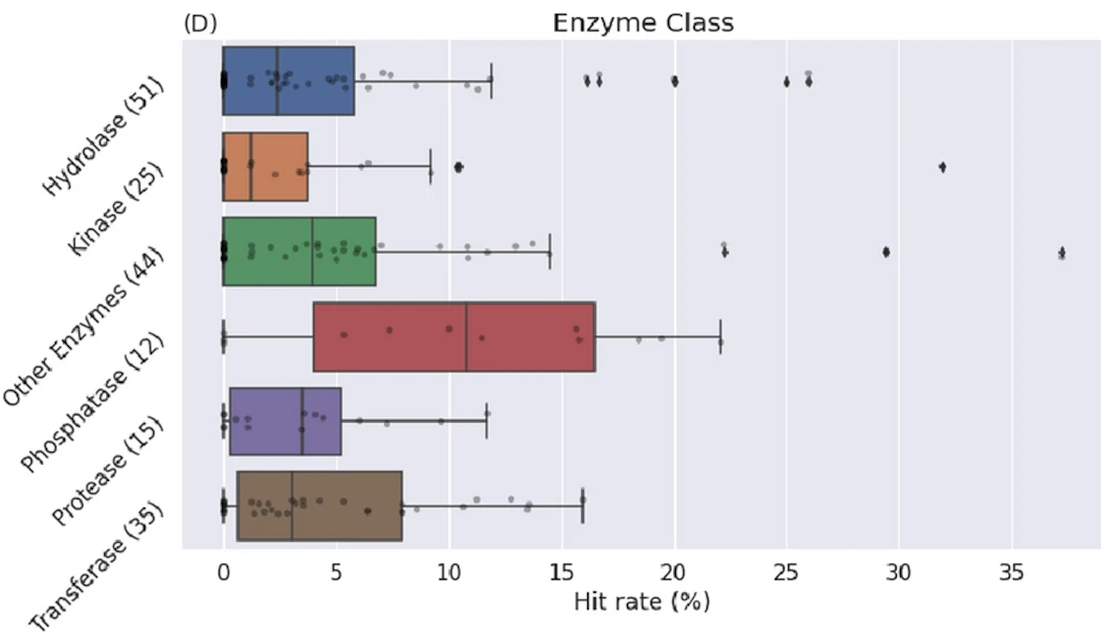
- PMID: 38565852
- PMCID: PMC10987645
- Full Text
- Atomwise, Inc.
- Atomwise press release
Guerrero L*, Ebrahim A*, Riley BT, Kim M, Huang Q, Finke AD, Keedy DA. Pushed to extremes: distinct effects of high temperature versus pressure on the structure of STEP. Communications Biology. 2024.
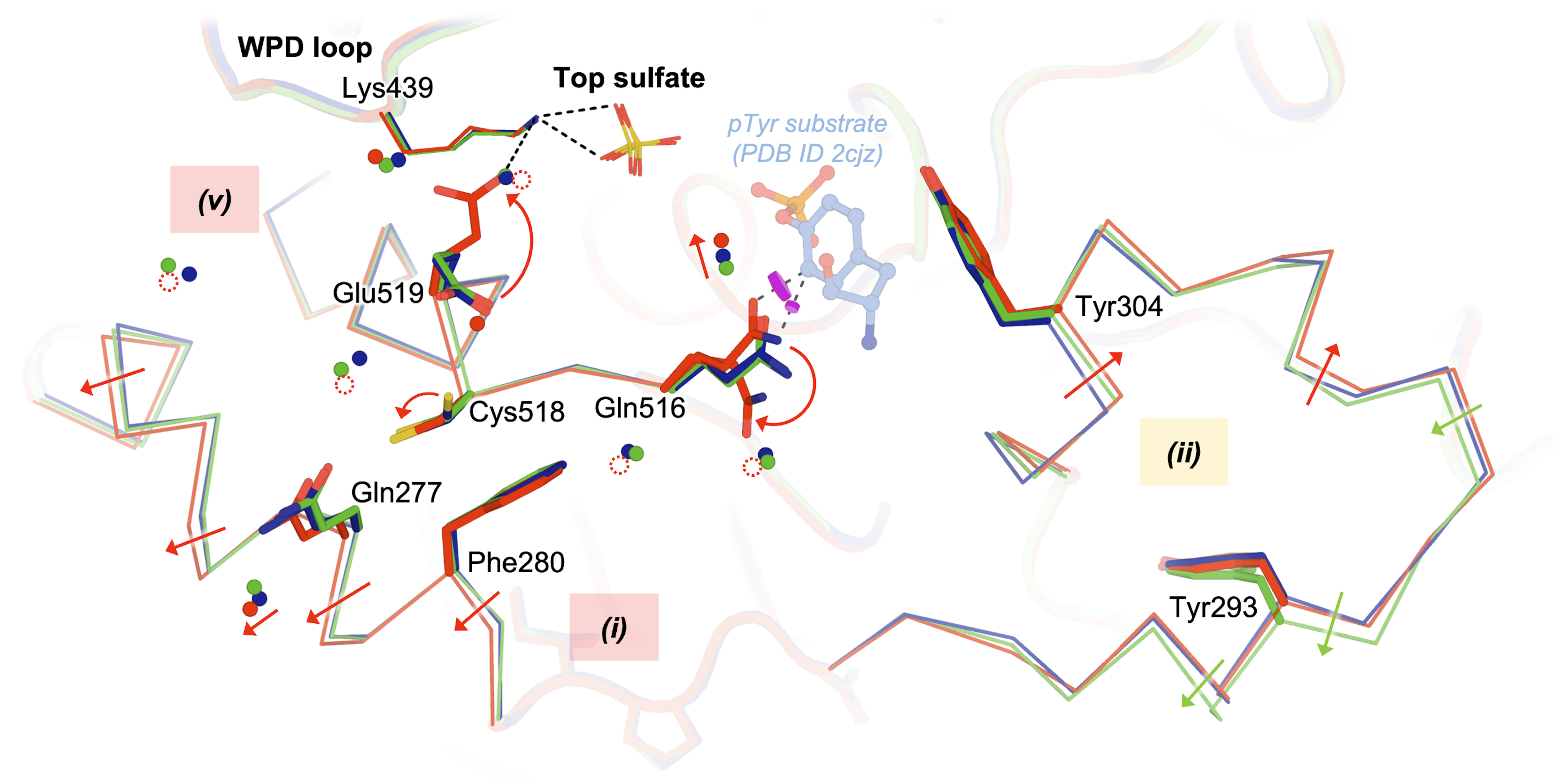
- PMID: 38216663
- PMCID: PMC10786866
- bioRxiv Preprint: 2023.05.02.538097
- Full Text
- Deposited Structures: 8SLS, 8SLT, 8SLU
- FlexX beamline at MacCHESS
- Springer Nature Communities Behind the Paper blog post
- CUNY ASRC news post
Sharma S, Mehlman (Skaist) T, Sagabala RS, Boivin B, Keedy DA. High-resolution double vision of the allosteric phosphatase PTP1B. Acta Cryst F. 2024.
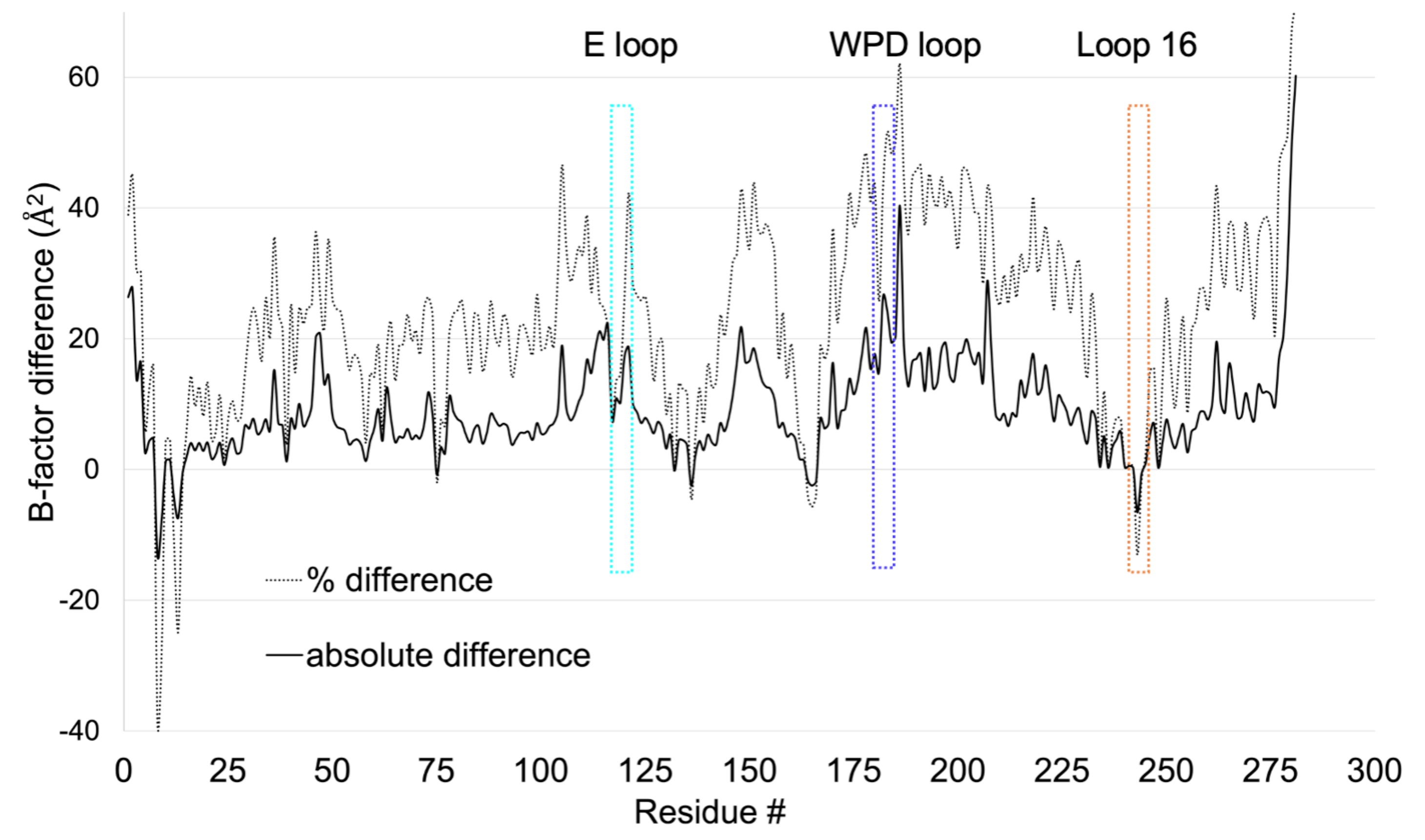
- PMID: 38133579
- PMCID: PMC10833341
- bioRxiv Preprint: 2023.09.12.557361
- Full Text
- Deposited Structure: 8U1E
- NYX beamline at NSLS-II
- Boivin lab at SUNY Poly
Mehlman (Skaist) T, Biel JT, Azeem SM, Nelson ER, Hossain S, Dunnett LE, Paterson NG, Douangamath A, Talon R, Axford D, Orins H, von Delft F, Keedy DA. Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B. eLife. 2023.
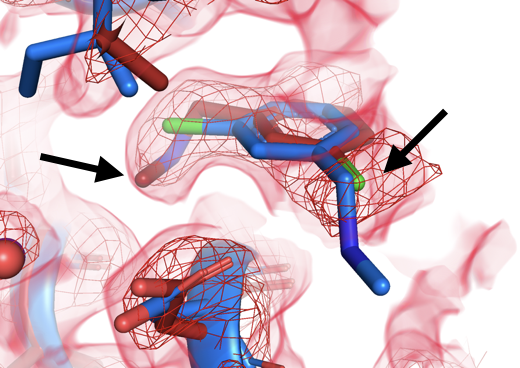
- PMID: 36881464
- PMCID: PMC9991056
- bioRxiv Preprint: 2022.11.02.514751
- Full Text
- Deposited Structures: 7FQM, 7FQN, 7FQO, 7FQP, 7FQQ, 7FQR, 7FQS, 7FQT, 7FQU, 7FQV, 7FQW, 7FQX, 7FQY, 7FQZ, 7FRF, 7FRG, 7FRH, 7FRI, 7FRJ, 7FRK, 7FRL, 7FRM, 7FRN, 7FRO, 7FRP, 7FRQ, 7FRR, 7FRE, 7FRS, 7FRT, 7FRU
- XChem - Fragment Screening at Diamond Light Source
- Datasets on Zenodo
- Twitter thread
- MiTeGen Newsletter
- Public reviews posted on bioRxiv preprint
- Practical Fragments - Crystallography heats up, it seems for the better
- Faculty Opinions Evaluation
- CUNY ASRC news post
Richardson JS, Moriarty NW, Keedy DA. Fitting Tip #23 - Alternate conformations always want to spread. Computational Crystallography Newsletter. 2023.
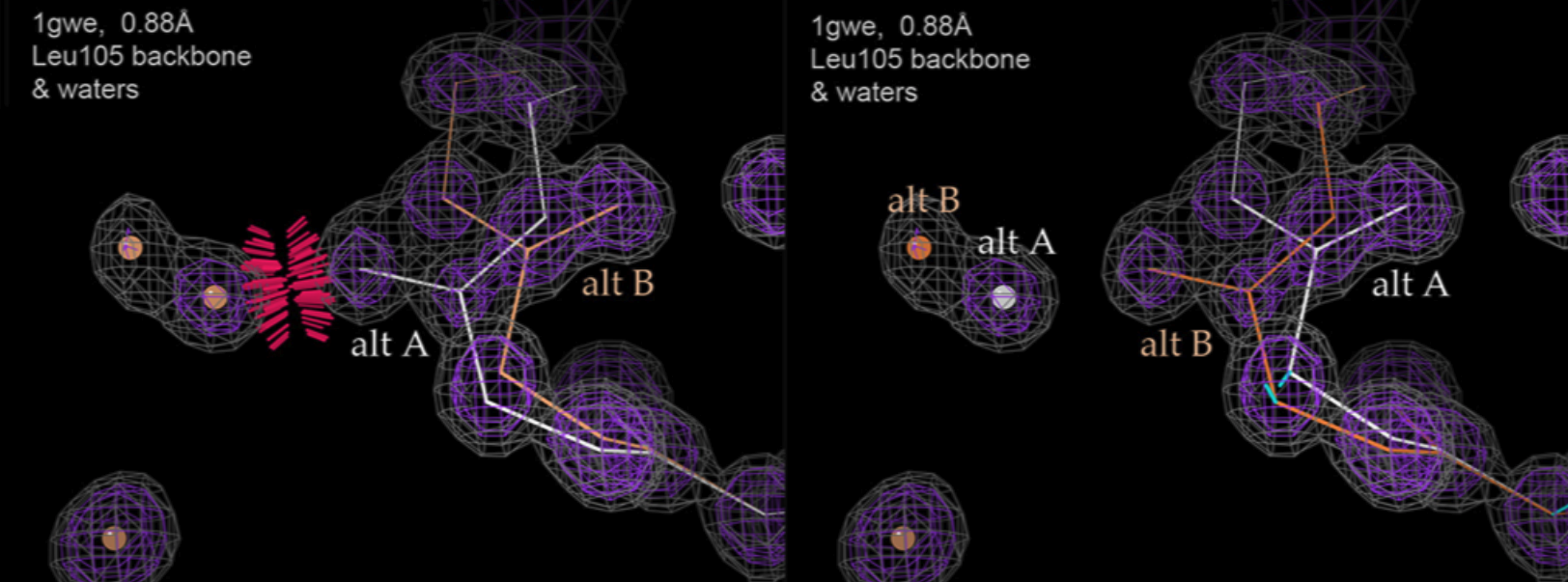
- PMID:
- Full Text
- PHENIX Computational Crystallography Newsletter
- Richardson Lab
Sharma S, Ebrahim A, Keedy DA. Room-temperature serial synchrotron crystallography of the human phosphatase PTP1B. Acta Cryst F. 2023.
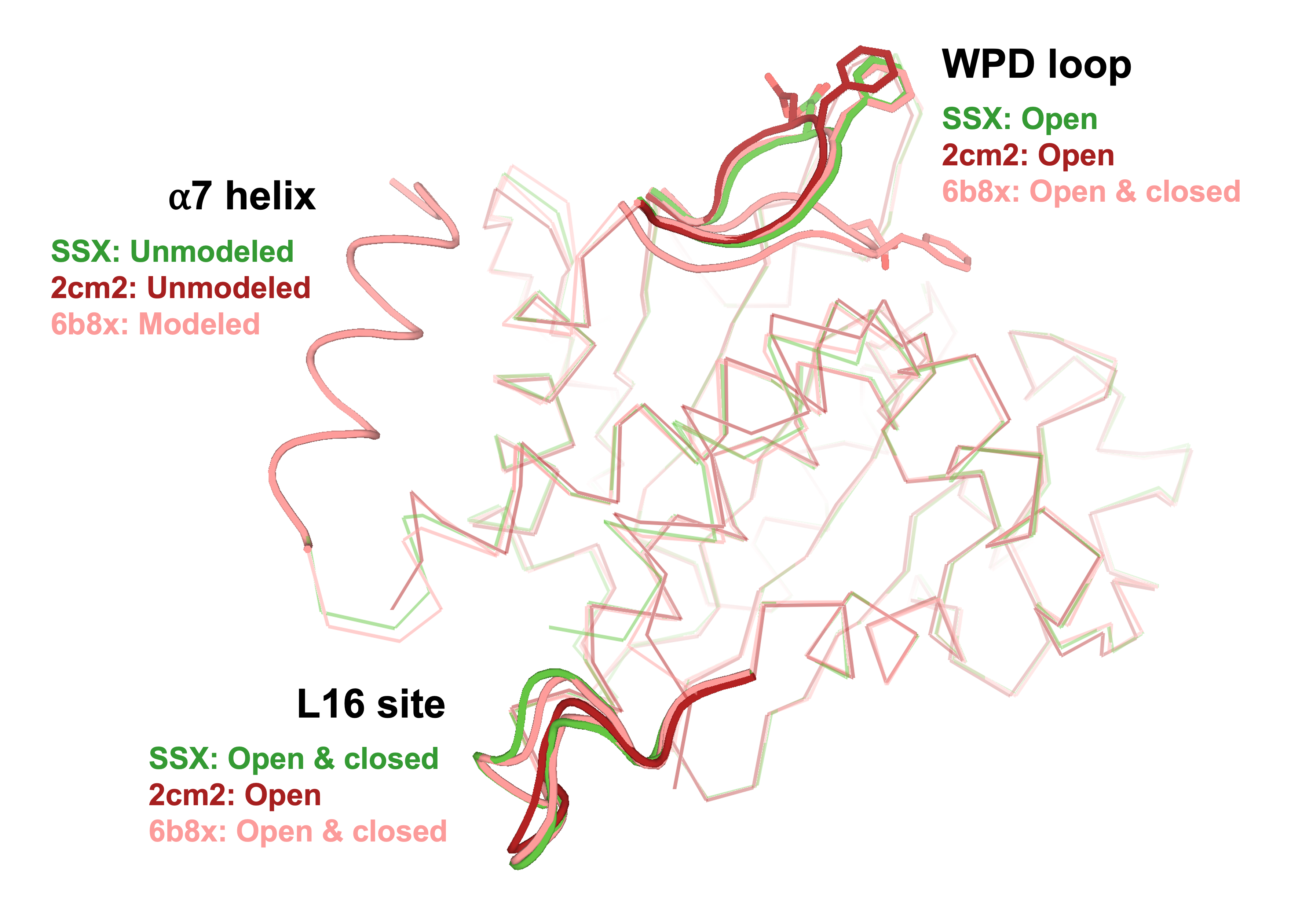
- PMID: 36598353
- PMCID: PMC9813971
- bioRxiv Preprint: 2022.07.28.501725
- Full Text
- Deposited Structure: 8DU7
- X-ray diffraction data at SBGrid Data Bank
- FlexX beamline at MacCHESS
- CCP4/APS School in Macromolecular Crystallography
- IUCr virtual thematic special issue on RT crystallography
Ebrahim A*, Riley BT*, Kumaran D, Andi B, Fuchs MR, McSweeney S, Keedy DA. The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (Mpro). IUCrJ. 2022.
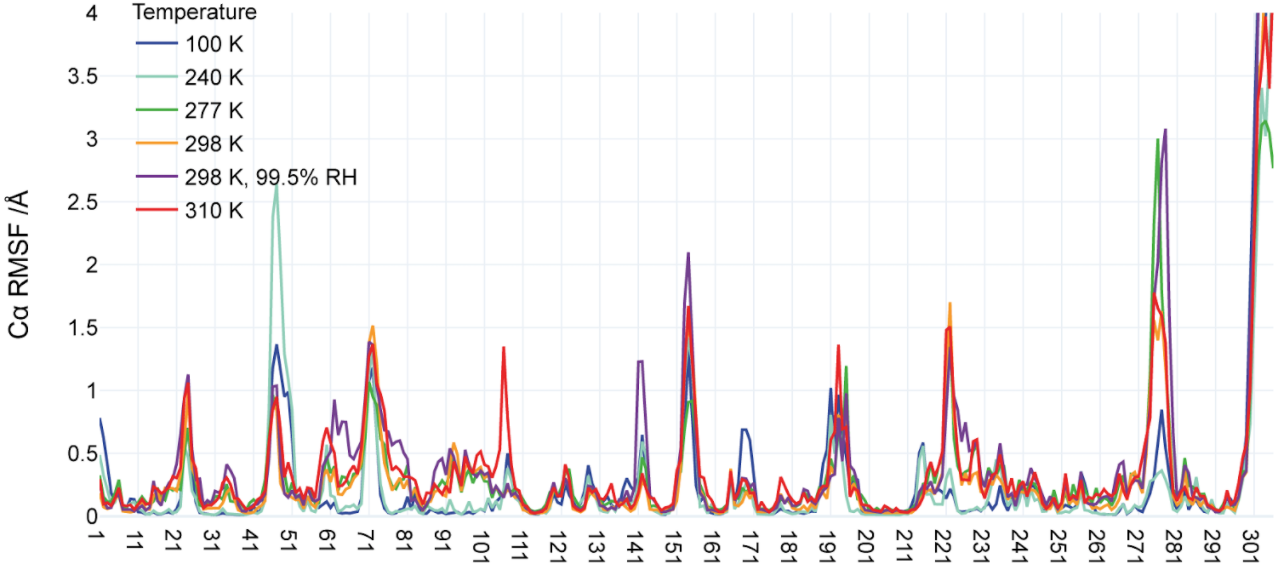
- PMID: 36071812
- PMCID: PMC9438506
- bioRxiv Preprint: 2021.05.03.437411
- Full Text
- Deposited Structures: 7MHF, 7MHG, 7MHH, 7MHI, 7MHJ, 7MHK, 7MHL, 7MHM, 7MHN, 7MHO, 7MHP, 7MHQ
- FMX beamline at NSLS-II
- Brookhaven National Lab press release
- Proteopedia article with interactive 3D graphics (click the links!)
- Kudos Research Showcase
- MiTeGen Newsletter
- IUCr virtual thematic special issue on RT crystallography
Riley BT, Wankowicz SA, de Oliveira SHP, van Zundert GCP, Hogan DW, Fraser JS, Keedy DA*, van den Bedem H* (* co-corresponding authors). qFit 3: Protein and ligand multiconformer modeling for X-ray crystallographic and single-particle cryo-EM density maps. Protein Science. 2021.
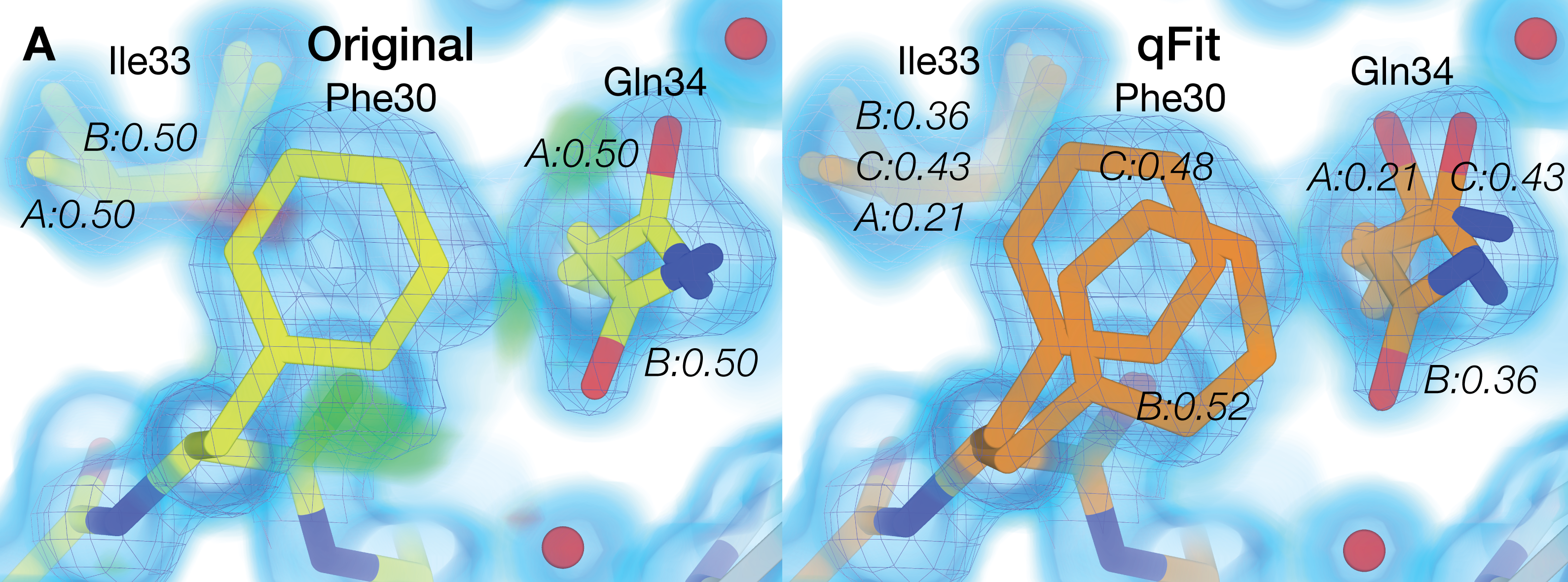
- PMID: 33210433
- PMCID: PMC7737783
- bioRxiv Preprint: 2020.09.03.280222
- Full Text
- qFit 3 Code on Github
- Henry van den Bedem
- James Fraser
- Schrodinger, Inc.
Keedy DA. Journey to the center of the protein: allostery from multitemperature multiconformer X-ray crystallography. Acta Cryst D. 2019.
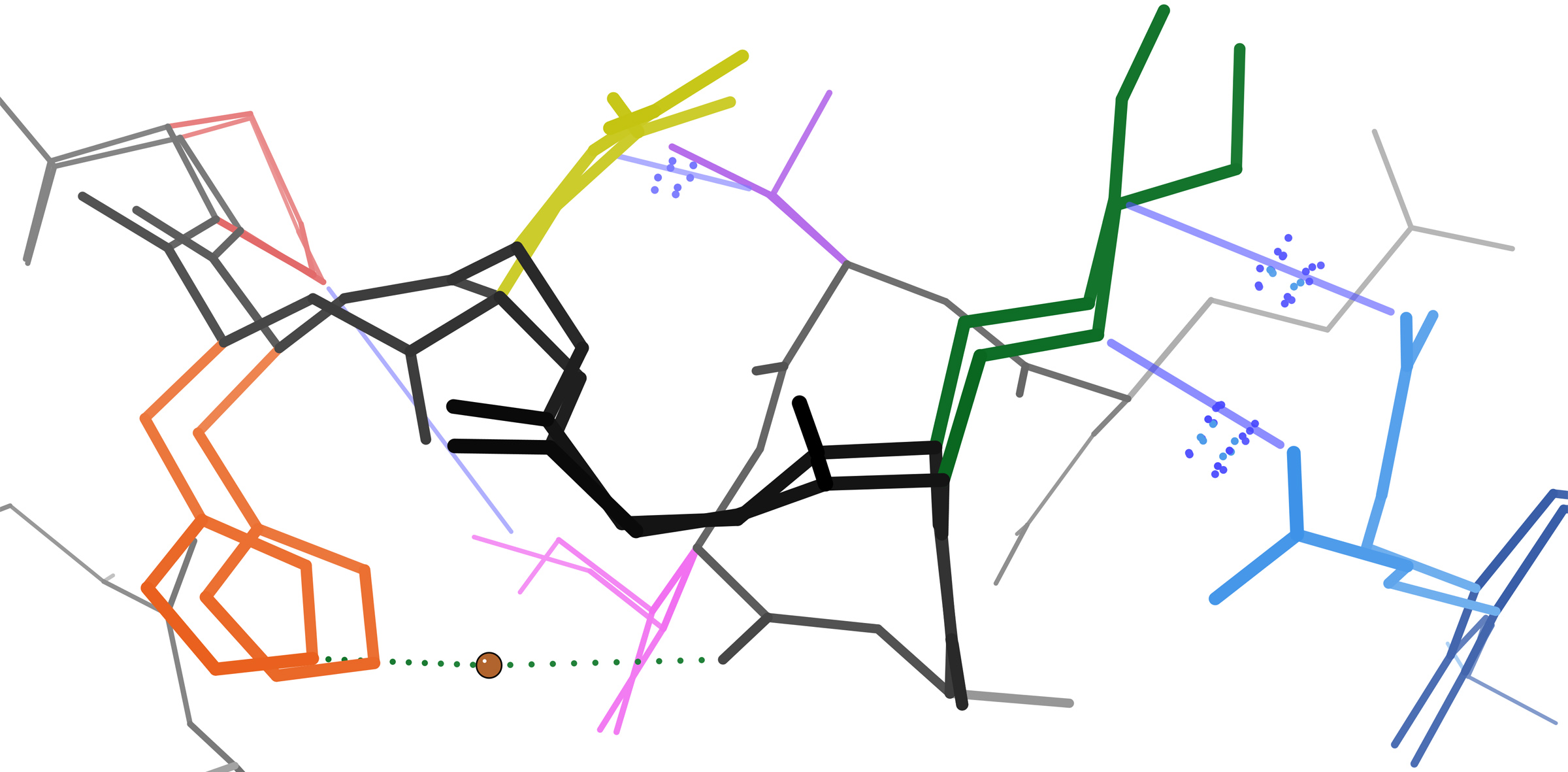
- PMID: 30821702
- PMCID: PMC6400254
- Full Text
- CCP4
van Zundert GCP*, Hudson BM*, Oliveira SHP, Keedy DA, Fonseca R, Heliou A, Suresh P, Borrelli K, Day T, Fraser JS, van den Bedem H. qFit-ligand reveals widespread conformational heterogeneity of drug-like molecules in X-ray electron density maps. Journal of Medicinal Chemistry. 2018.
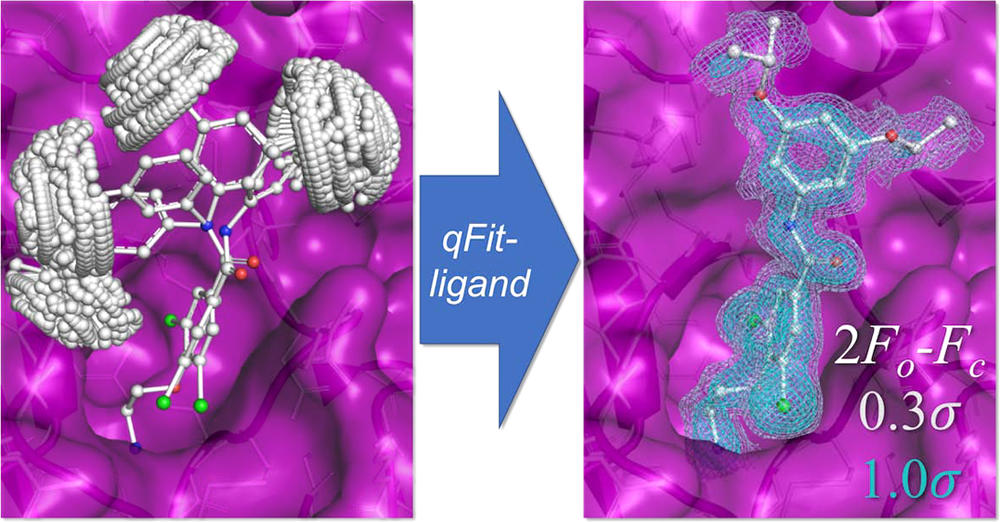
- PMID: 30457858
- PMCID: PMC6820680
- bioRxiv Preprint: 253419
- Full Text
- Deposited Structures: 6DMG, 6DMH, 6DMI, 6DMJ, 6DMK, 6DML
- qFit-ligand Code on Github
- In the Pipeline-More Than One, And Maybe More Than That
- Practical Fragments - (Not) getting misled by crystal structures - part 5 – conformational heterogeneity
- James Fraser
- Henry van den Bedem
- Schrodinger, Inc.
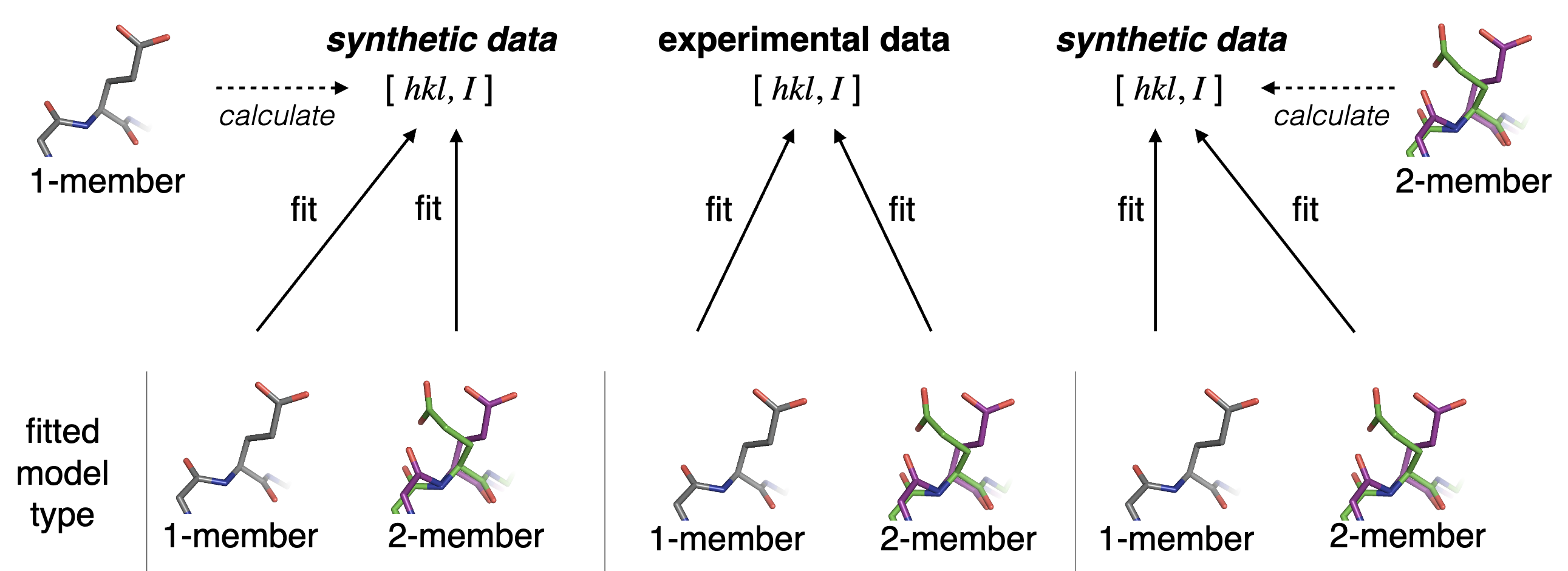
Babcock NS, Keedy DA, Fraser JS, Sivak DA. Model selection for biological crystallography. bioRxiv preprint. 2018.
- PMID:
- bioRxiv Preprint: 448795
- Full Text
- David Sivak
- James Fraser
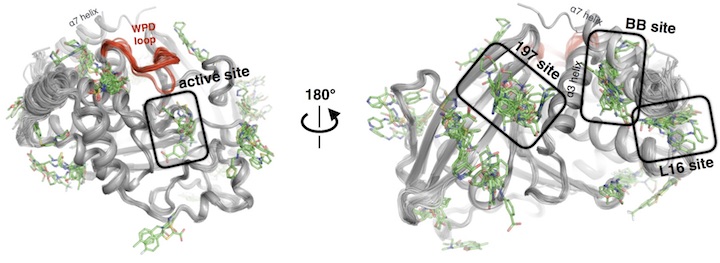
Keedy DA*, Hill ZB*, Biel JT, Kang E, Rettenmaier TJ, Brandao-Neto J, Pearce NM, von Delft F, Wells JA, Fraser JS. An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering. eLife. 2018.
- PMID: 29877794
- PMCID: PMC6039181
- bioRxiv Preprint: 218966
- Full Text
- Deposited Structures: 6B90, 6B8E, 6B8T, 6B8X, 6B8Z, 6BAI, 6B95, 5QDE, 5QDF, 5QDG, 5QDH, 5QDI, 5QDJ, 5QDK, 5QDL, 5QDM, 5QDN, 5QDO, 5QDP, 5QDQ, 5QDR, 5QDS, 5QDT, 5QDU, 5QDV, 5QDW, 5QDX, 5QDY, 5QDZ, 5QE0, 5QE1, 5QE2, 5QE3, 5QE4, 5QE5, 5QE6, 5QE7, 5QE8, 5QE9, 5QEA, 5QEB, 5QEC, 5QED, 5QEE, 5QEF, 5QEG, 5QEH, 5QEI, 5QEJ, 5QEK, 5QEL, 5QEM, 5QEN, 5QEO, 5QEP, 5QEQ, 5QER, 5QES, 5QET, 5QEU, 5QEV, 5QEW, 5QEX, 5QEY, 5QEZ, 5QF0, 5QF1, 5QF2, 5QF3, 5QF4, 5QF5, 5QF6, 5QF7, 5QF8, 5QF9, 5QFA, 5QFB, 5QFC, 5QFD, 5QFE, 5QFF, 5QFG, 5QFH, 5QFI, 5QFJ, 5QFK, 5QFL, 5QFM, 5QFN, 5QFO, 5QFP, 5QFQ, 5QFR, 5QFS, 5QFT, 5QFU, 5QFV, 5QFW, 5QFX, 5QFY, 5QFZ, 5QG0, 5QG1, 5QG2, 5QG3, 5QG4, 5QG5, 5QG6, 5QG7, 5QG8, 5QG9, 5QGA, 5QGB, 5QGC, 5QGD, 5QGE, 5QGF
- XChem - Fragment Screening at Diamond Light Source
- PanDDA code on Bitbucket
- James Fraser
- Jim Wells
- Datasets on Zenodo
- Paper Submission Celebration Photo
- Oxford Protein Informatics Group (OPIG) Journal Club blog
- Reviewed in Diamond Light Source Annual Review 2019
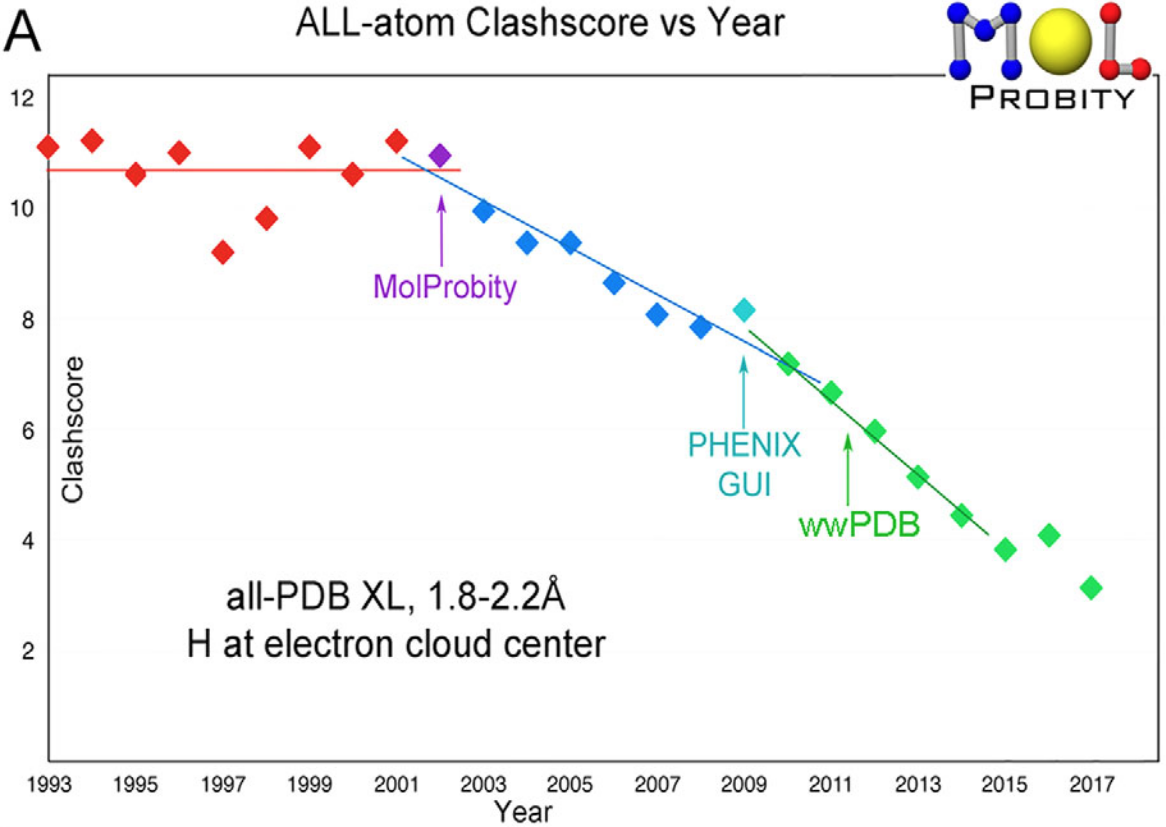
Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, Jain S, Lewis SM, Arendall WB 3rd, Snoeyink J, Adams PD, Lovell SC, Richardson JS, Richardson DC. MolProbity: More and better reference data for improved all-atom structure validation. Protein Science. 2018.
- PMID: 29067766
- PMCID: PMC5734394
- Full Text
- MolProbity model validation server
- Jane and David Richardson
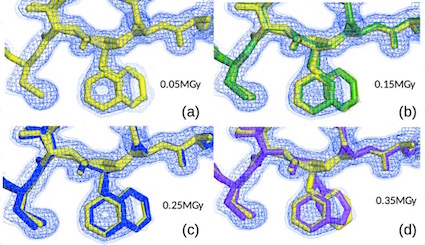
Russi S, González A, Kenner LR, Keedy DA, Fraser JS, van den Bedem H. Conformational variation of proteins at room temperature is not dominated by radiation damage. Journal of Synchrotron Radiation. 2017.
- PMID: 28009548
- PMCID: PMC5182021
- Full Text
- Henry van den Bedem
- James Fraser
Cimermancic P, Weinkam P, Rettenmaier TJ, Bichmann L, Keedy DA, Woldeyes RA, Schneidmann D, Demerdash ONA, Mitchell JC, Wells JA, Fraser JS, Sali A. CryptoSite: Expanding the druggable proteome by characterization and prediction of cryptic binding sites. Journal of Molecular Biology. 2016
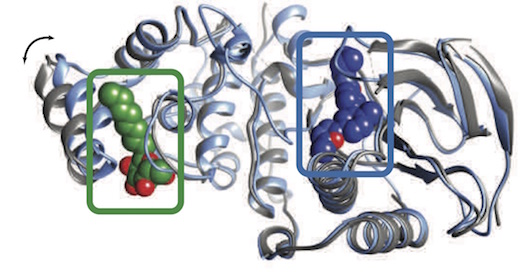
- PMID: 26854760
- PMCID: PMC4794384
- Full Text
- Paper Submission Celebration Photo
- CryptoSite
- James Fraser
- In the pipeline
- F1000 Evaluation
Keedy DA. Conformational and connotational heterogeneity: A surprising relationship between protein structural flexibility and puns. Proteins: Structure, Function, and Bioinformatics. 2015.
Keedy DA, Fraser JS, van den Bedem H. Exposing hidden alternative backbone conformations in X-ray crystallography using qFit. PLOS Computational Biology. 2015.
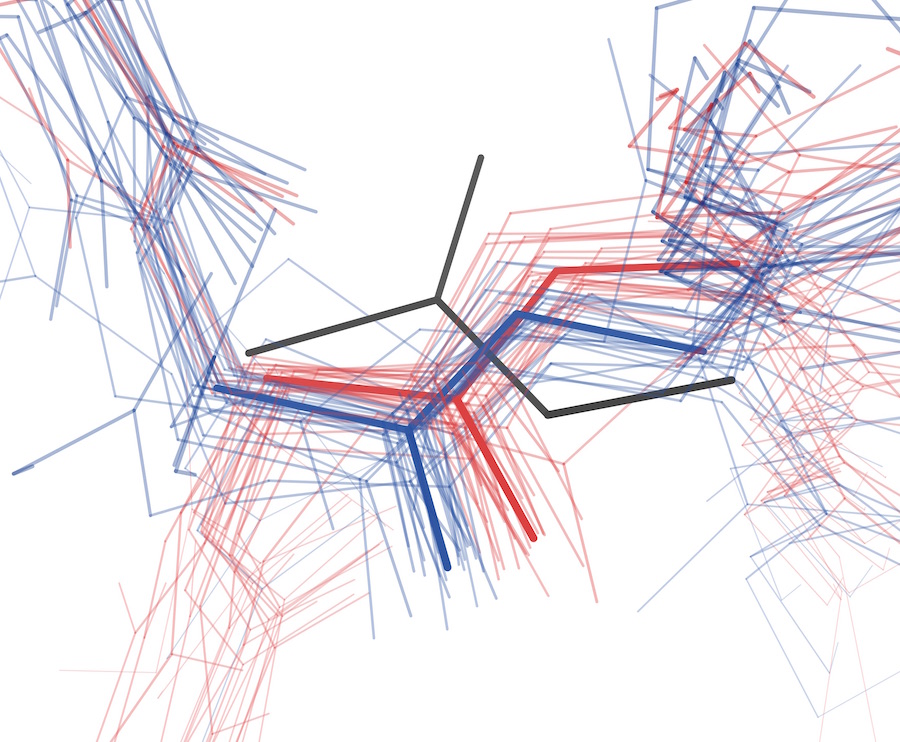
- PMID: 26506617
- PMCID: PMC4624436
- bioRxiv Preprint: 018879
- Full Text
- Henry van den Bedem
- James Fraser
- qFit webserver
- qFit code
- Coordinates for peptide flip clusters
- Paper Submission Celebration Photo
Keedy DA*, Kenner LR*, Warkentin M*, Woldeyes RA*, Hopkins JB, Thompson MC, Brewster AS, Van Benschoten AH, Baxter EL, Uervirojnangkoorn M, McPhillips SE, Song J, Alonso-Mori R, Holton JM, Weis WI, Brunger AT, Soltis SM, Lemke H, Gonzalez A, Sauter NK, Cohen AE, van den Bedem H, Thorne RE, Fraser JS. Mapping the Conformational Landscape of a Dynamic Enzyme by Multitemperature and XFEL Crystallography. eLife. 2015.
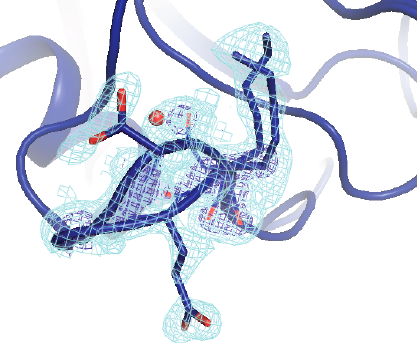
- PMID: 26422513
- PMCID: PMC4721965
- bioRxiv Preprint: 016733
- Full Text
- Online Dataset: doi:10.15785/SBGRID/68
- Deposited Structures: 4YUG, 4YUH, 4YUI, 4YUJ, 4YUK, 4YUL, 4YUM, 4YUN, 4YUO, 4YUP
- Paper Submission Celebration Photo
- Protein Harlem Shake on YouTube
- LCLS XPP Beamline
- Henry van den Bedem
- Rob Thorne
- James Fraser
Keedy DA, van den Bedem H, Sivak DA, Petsko GA, Ringe D, Wilson MA, Fraser JS. Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR. Structure. 2014.
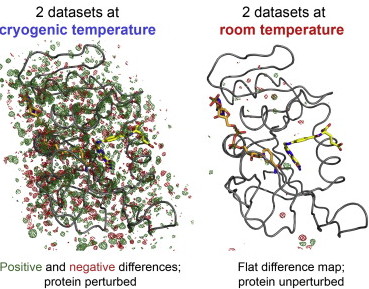
- PMID: 24882744
- PMCID: PMC4082491
- Full Text
- Deposited Structures: 4PSS, 4PST, 4PSY, 4PSZ, 4PTJ, 4P3Q, 4P3R, 4PTH
- Mark Wilson
- Greg Petsko / Dagmar Ringe
- James Fraser
- Paper Submission Celebration Photo
- F1000 Evaluation
Keedy DA. Using Protein-Likeness to Validate Conformational Alternatives. PhD Thesis. 2012.

- PMID:
- Full Text
- Jane and Dave Richardson
- Bruce Donald
- Duke Biochemistry